
5.1.1. CMIP6 biases in SPEI6 drought simulations over the Mediterranean region#
Production date: 20-09-2025
Produced by: CMCC foundation - Euro-Mediterranean Center on Climate Change. Albert Martinez Boti and Lorenzo Sangelantoni.
🌍 Use case: Investigating drought biases to inform water management#
❓ Quality assessment question#
How well do CMIP6 models simulate drought compared to signals derived from ERA5?
The Standardized Precipitation-Evapotranspiration Index (SPEI) [1] is a widely used drought indicator that integrates precipitation and potential evapotranspiration, providing a more complete measure of water deficits than precipitation alone. Its multiscalar formulation allows the evaluation of different types of drought [2], from short-term agricultural or soil moisture deficits to long-term hydrological or ecological events. Being standardized, SPEI is dimensionless, facilitating comparisons across regions and climates. By capturing variations in frequency, duration, and severity, it effectively reflects the complex nature of drought, making it a robust and physically meaningful proxy for drought assessment in the context of climate change [3].
Understanding SPEI is particularly relevant for the water management sector. By capturing multiple types of drought SPEI serves as a versatile indicator for various users. Climate projections using SPEI enable planners to anticipate future water deficits, guide allocation, and design effective strategies for reservoir management, irrigation, and drought preparedness, helping to reduce socio-economic and ecological impacts [4][5].
This assessment evaluates SPEI6 (six-month accumulation) across the full ensemble of CMIP6 models available through the Copernicus Climate Data Store (CDS). Model outputs are compared against SPEI6 derived from ERA5 reanalysis, which serves as the reference product. The standard reference period is 1971–2010. We specifically evaluate whether changes in the 1991–2020 period relative to the 1961–1990 baseline are accurately represented by CMIP6 models and the trend acroos the whole 1961-2020 period for the Mediterranean domain.
📢 Quality assessment statement#
These are the key outcomes of this assessment
CMIP6 ensemble median captures the general intensification of drought over the Mediterranean seen in ERA5, though the magnitude of the change is underestimated, particularly over eastern Spain, southern France, and much of Turkey.
Individual models do not reproduce the same spatial pattern, highlighting the importance of using large ensembles rather than relying on a single model for planning purposes.
Despite inherent biases, the considered subset of CMIP6 models provides a foundation for understanding the evolution of past drought conditions. These results support the use of SPEI to anticipate water deficits and guide drought preparedness, increasing confidence (though not ensuring accuracy) when assessing future trends.
Regions with high inter-model spread, such as North Africa, Turkey, and the eastern Mediterranean, may be associated with future higher uncertainty, suggesting that adaptation strategies in these areas should consider a wider range of possible futures.
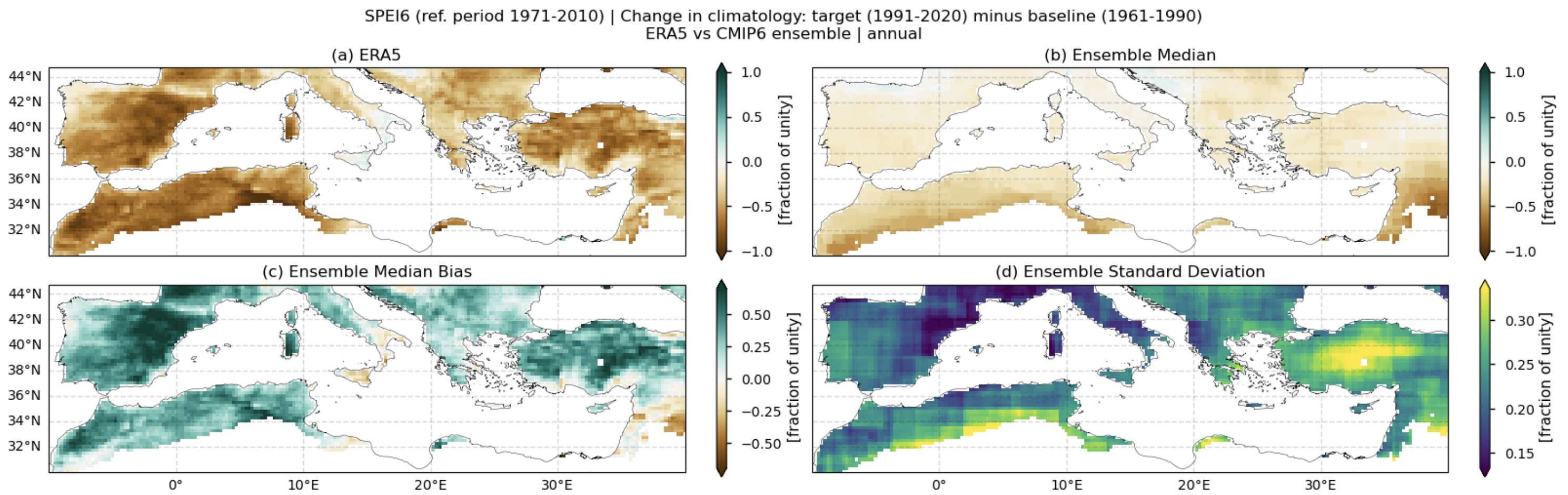
Fig. 5.1.1.1 Spatial patterns of SPEI6 change over the Mediterranean region. Panels show (a) ERA5 reference, (b) CMIP6 ensemble median (median of the selected subset of models at each grid cell), (c) ensemble median bias relative to ERA5, and (d) ensemble spread (standard deviation across the selected models). SPEI6 is standardised over the reference period 1971–2010. SPEI6 change is calculated at each grid point as the difference between the monthly climatologies of the target period (1991–2020) and the baseline period (1961–1990), and then averaged to obtain the annual mean.#
📋 Methodology#
The Standardized Precipitation-Evapotranspiration Index accumulated to six months (SPEI6) has been calculated for this assessment — through the use of xclim — as a proxy to evaluate drought conditions for a subset of CMIP6 models, which are evaluated against SPEI6 derived from ERA5 reanalysis. The study covers the Mediterranean domain, following IPCC-AR6 regional definitions [6], and results include spatial maps of SPEI6 changes for ERA5, CMIP6, and the biases of these changes. Additionally, time series showing the temporal evolution of drought across the domain are calculated, comparing trends from the models with ERA5 over the historical period considered within this assessment (1961–2020).
Potential evapotranspiration (PET) is calculated with the Hargreaves method [7], which requires only maximum and minimum temperatures (and optionally mean temperature), providing a computationally efficient yet robust estimate (e.g., [8],). The accumulated series of precipitation minus PET is standardized for the reference period defined by the C3S Atlas (1971–2010).
Five key concepts are defined:
Reference period: The period used to standardize SPEI values, here 1971–2010, following the C3S Atlas.
Baseline period: The period against which changes are calculated, here 1961–1990.
Target period: The period for which changes are evaluated, here 1991–2020.
Historical period: The entire period considered in this assessment, here 1961–2020.
Change assessment: Differences in SPEI6 between the target period and the baseline, representing anomalies in drought conditions.
The analysis and results follow the next outline:
1. Parameters, requests and functions definition
📈 Analysis and results#
1. Parameters, requests and functions definition#
1.1. Import packages#
1.2. Define Parameters#
In the “Define Parameters” section, various customisable options for the notebook are specified:
The historical period can be adjusted by modifying
historical_slice; the default range is 1961-2020.The reference period can be adjusted by modifying
reference_slice; the default range is 1971–2010, following the C3S Atlas.The baseline period can be adjusted by modifying
baseline_slice; the default range is 1961-1990.The target period can be adjusted by modifying
target_slice; the default range is 1991-2020.The
interpolation_methodparameter allows selecting the interpolation method when regridding is performed.collection_idis not customisable for this assessment and is set to ‘CMIP6’. Expert users can use this notebook as a guide to create similar analyses for other model sets (such as CORDEX).The
areaparameter specifies the geographical domain of interest. By default, the Mediterranean domain is used, following IPCC-AR6 regional definitions [6]The
time_aggallows selecting the time aggregation for the analysis. Options include: annual, DJF, MAM, JJA, SON, Jan, Feb, Mar, Apr, May, Jun, Jul, Aug, Sep, Oct, Nov, Dec.SPEI accumulation window (
window): Defines the accumulation period in months for the SPEI calculation. Common options include 3, 6, or 12 months, depending on whether short-term or long-term droughts are of interest. In this assessment, a six-month accumulation is used.The
chunksselection allows the user to define if dividing into chunks when downloading the data on their local machine. Although it does not significantly affect the analysis, it is recommended to keep the default value for optimal performance.
1.3. Define models#
The following climate analyses are performed using CMIP6 models that provide both the historical and SSP5-8.5 experiments, as well as maximum, minimum and mean temperatures and precipitation, within the Climate Data Store (CDS).
1.4. Define ERA5 request#
Within this notebook, ERA5 serves as the reference product. In this section, we set the required parameters for the cds-api data-request of ERA5.
1.5. Define model requests#
In this section we set the required parameters for the cds-api data-request.
1.6. Functions to cache#
In this section, functions that will be executed in the caching phase are defined. Caching is the process of storing copies of files in a temporary storage location, so that they can be accessed more quickly. This process also checks if the user has already downloaded a file, avoiding redundant downloads.
Description of the main functions:
compute_spei: Uses the xclim package to calculate the Standardized Precipitation-Evapotranspiration Index (SPEI). Thereference_sliceargument specifies the reference period used for standardization, while thewindowargument defines the number of months over which precipitation and evapotranspiration are accumulated (6 months in this assessment).get_mask_below_03: Generates a mask identifying grid points where precipitation is below 0.3 mm/day, allowing filtering of extremely dry regions.
2. Downloading and processing#
2.1. Download and transform ERA5#
In this section, the download.download_and_transform function from the ‘c3s_eqc_automatic_quality_control’ package is used to retrieve ERA5 hourly reference data, resample it to daily frequency, compute daily potential evapotranspiration, calculate the daily water balance, and derive the SPEI (SPEI6 for this assessment). Results are cached to avoid redundant downloads and repeated processing.
2.2. Download and transform models#
In this section, the download.download_and_transform function from the ‘c3s_eqc_automatic_quality_control’ package is used to download daily data from CMIP6 models, compute daily potential evapotranspiration, calculate the daily water balance, and derive the SPEI (SPEI6 for this assessment). SPEI6 is computed on each model’s native grid and also on the ERA5 grid to enable bias assessment. When regridding is required, it is performed for each essential climate variable before the index calculation to preserve standardisation. Results are cached to avoid redundant downloads and repeated processing.
model='access_cm2'
model='canesm5'
model='cmcc_esm2'
model='cnrm_cm6_1'
model='cnrm_cm6_1_hr'
model='cnrm_esm2_1'
model='ec_earth3_cc'
model='gfdl_esm4'
model='inm_cm4_8'
model='inm_cm5_0'
model='kace_1_0_g'
model='miroc6'
model='miroc_es2l'
model='mpi_esm1_2_lr'
model='mri_esm2_0'
model='nesm3'
model='noresm2_mm'
2.3. Apply land-sea mask and change some attributes#
This section changes some attributes and applies a land–sea mask to all models, as well as to ERA5. A bare-earth (barrean) mask is also applied to ensure meaningful results in arid regions, as in the C3S atlas. In particular, we follow the methodology available in their Github repository. While their implementation of the mask also considers factors such as snow depth and vegetation cover, here we only apply the filter based on annual mean precipitation for 1991–2020 being less than 0.3 mm day⁻¹. This simplification is justified because, in the region under consideration, the bare-earth mask obtained with the full set of filters closely matches the one derived from the precipitation threshold alone.
Note: ds_interpolated contains data from the models regridded to the ERA5 grid. model_datasets contain the same data on the original grid of each model. Regridding is performed for every essential climate variable prior to the index calculation in order to avoid compromising standardisation.
100%|██████████| 1/1 [00:00<00:00, 44.33it/s]
3. Plot and describe results#
This section will display the following results:
SPEI6 change maps comparing ERA5 and the ensemble median (defined as the median of the change values for the selected subset of models at each grid cell). The layout includes ERA5, the ensemble median, the ensemble median bias, and the ensemble spread (calculated as the standard deviation of the change across the selected subset of models).
SPEI6 change maps for each individual model.
Bias maps of the SPEI6 change for each model.
Time series of SPEI6 change (averaged over the Mediterranean region).
Note: SPEI6 change is calculated at each grid point as the arithmetic difference between climatologies in the target period (1991–2020) and the baseline period (1961–1990).
3.1. Define plotting functions#
The functions presented here are used to calculate SPEI6 changes and generate corresponding layout plots. Three types of layout can be displayed, depending on the plotting function:
Reference and ensemble summary: Includes the ERA5 reference product, the ensemble median, the bias of the ensemble median, and the ensemble spread. This is generated using
plot_ensemble().Individual models: Displays all models individually using
plot_models().Model biases: Displays the bias of each model relative to the reference, also using
plot_models().
Calculation of SPEI6 change:
compute_spei_change()calculates SPEI6 change at each grid point as the arithmetic difference between monthly climatologies of the target period (1991–2020) and the baseline period (1961–1990). It then aggregates the change according to the user-specifiedtime_agg(annual, seasonal, or monthly).compute_change_4timeseries()computes SPEI6 anomalies relative to the baseline climatology for each month of the timeseries and aggregates them according to thetime_aggparameter. This provides monthly, seasonal, or annual-level information depending on the user’s selection.
3.2. Plot ensemble maps#
In this section, we invoke the plot_ensemble() function to visualise the SPEI6 change for the following: (a) the reference ERA5 product, (b) the ensemble median (defined as the median of the change values for the selected subset of models at each grid cell), (c) the bias of the ensemble median, and (d) the ensemble spread (calculated as the standard deviation of the change across the selected subset of models).
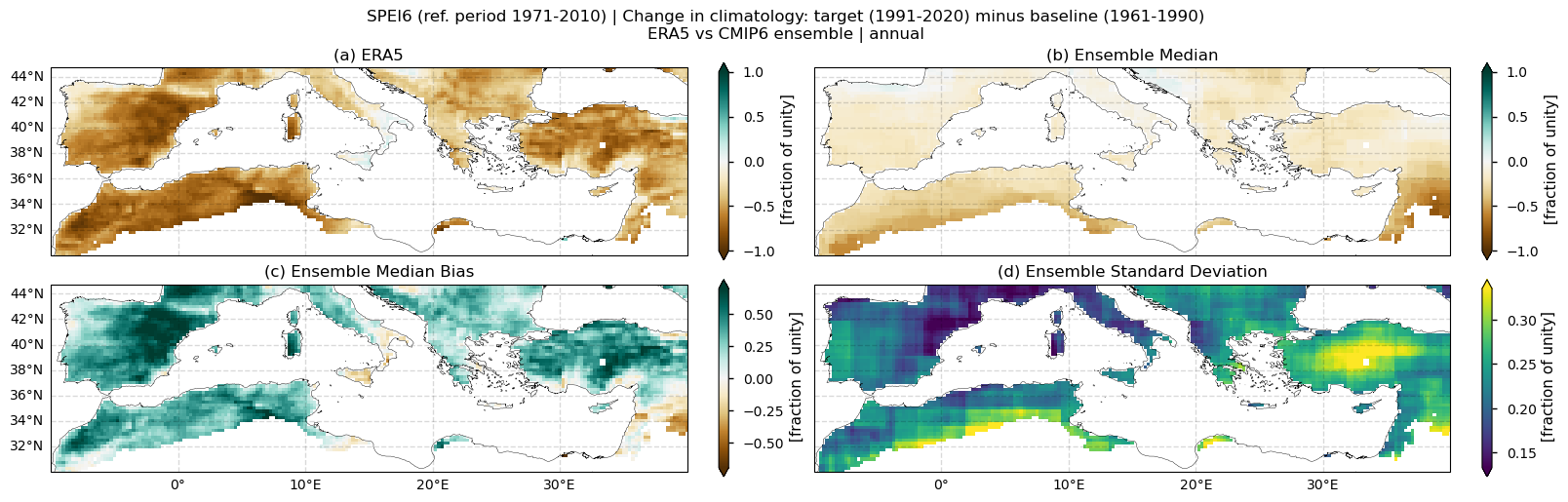
Fig 1. Spatial patterns of SPEI6 change over the Mediterranean region. Panels show (a) ERA5 reference, (b) CMIP6 ensemble median (median of the selected subset of models at each grid cell), (c) ensemble median bias relative to ERA5, and (d) ensemble spread (standard deviation across the selected models). SPEI6 is standardised over the reference period 1971–2010. SPEI6 change is calculated at each grid point as the difference between the monthly climatologies of the target period (1991–2020) and the baseline period (1961–1990), and then averaged to obtain the annual mean.
3.3. Plot model maps#
In this section, we invoke the plot_models() function to visualise the SPEI6 change of every model individually. Note that the model data used in this section maintains its original grid.
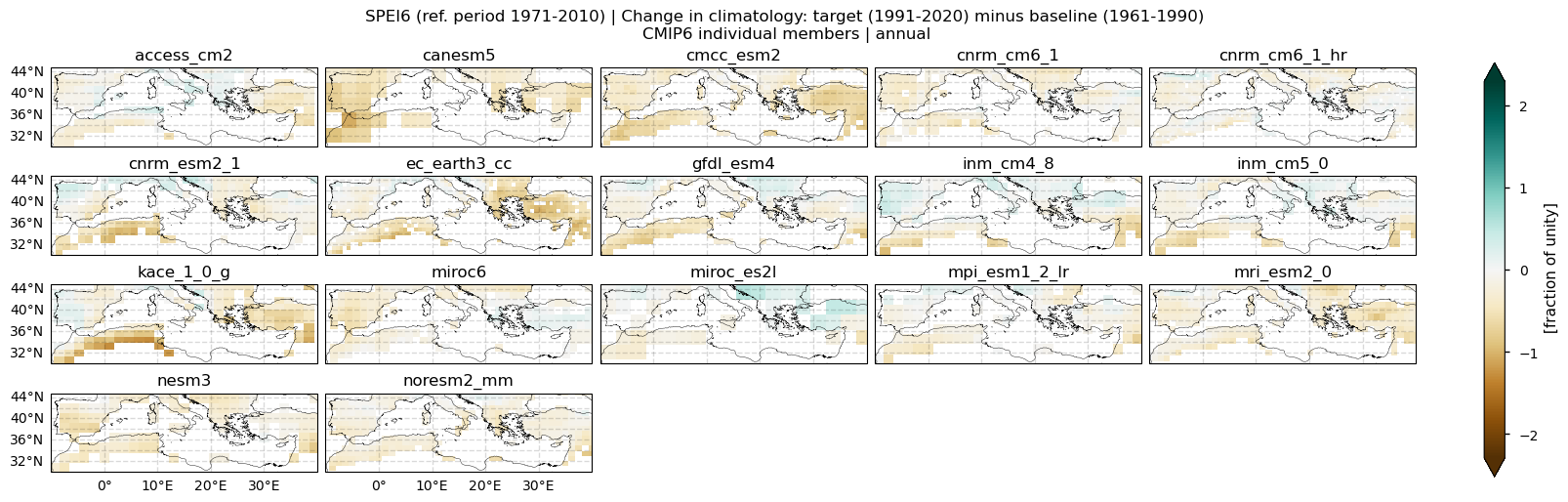
Fig 2. Spatial patterns of SPEI6 change for each individual CMIP6 model over the Mediterranean region. SPEI6 is standardised over the reference period 1971–2010. For each model, SPEI6 change is calculated at each grid point as the difference between the monthly climatologies of the target period (1991–2020) and the baseline period (1961–1990), and then averaged to obtain the annual mean.
3.4. Plot bias maps#
In this section, we invoke the plot_models() function to visualise the bias of the vSPEI6 change for every model individually. Note that the model data used in this section has previously been interpolated to the ERA5 grid. Regridding is performed for each essential climate variable before the index calculation to preserve standardisation.
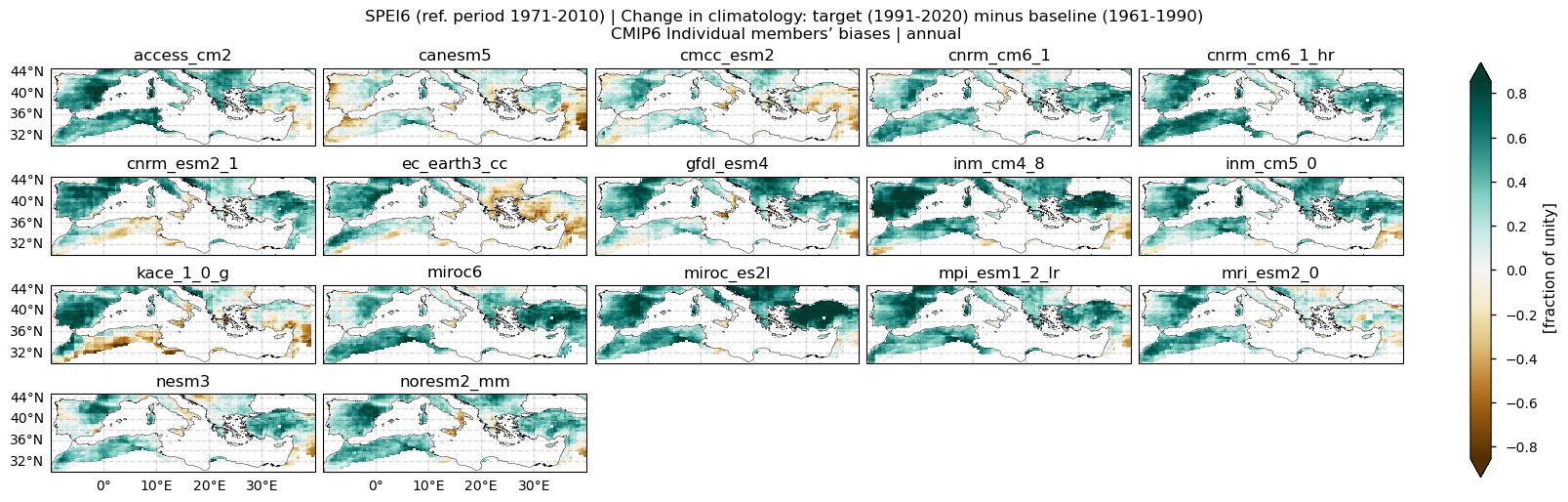
Fig 3. Bias of SPEI6 change for each individual CMIP6 model relative to ERA5 over the Mediterranean region. SPEI6 is standardised over the reference period 1971–2010. For each model and ERA5, SPEI6 change is calculated at each grid point as the difference between the monthly climatologies of the target period (1991–2020) and the baseline period (1961–1990), and then averaged to obtain the annual mean.
3.5. Timeseries#
This section examines the time series of SPEI6 anomalies relative to the baseline climatology. We first use compute_change_4timeseries(), which calculates SPEI6 anomalies for each month of the timeseries (relative to the baseline climatology of each month) and aggregates them according to the time_agg parameter. Spatially averaged values are then obtained. Light grey shading highlights the baseline period, while darker grey shading marks the target period. The analysis compares the CMIP6 ensemble median, ERA5, and individual models, displaying trends for both ERA5 and the ensemble median. Additionally, it shows the ensemble spread and the slope values of the trends for ERA5 and the CMIP6 ensemble median.
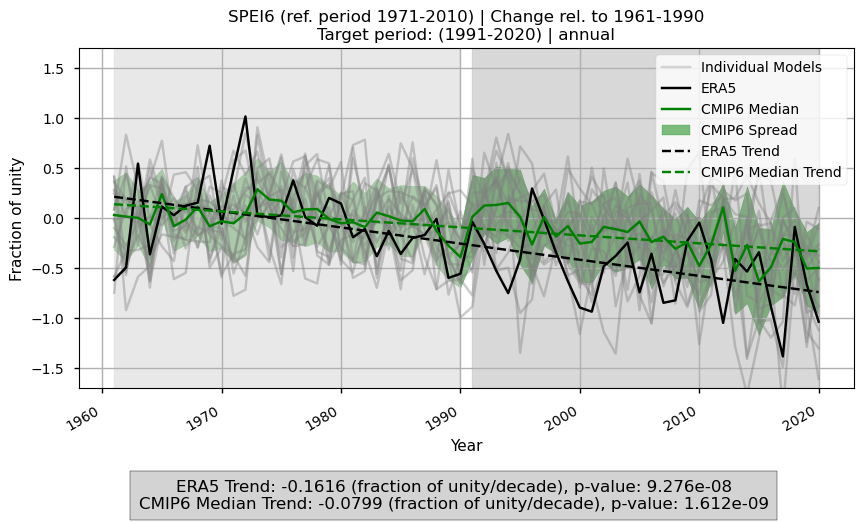
Fig 4. Time series of SPEI6 anomalies over the Mediterranean region, calculated relative to the baseline climatology. Monthly anomalies are aggregated according to the specified timescale and then spatially averaged over the region. Light grey shading indicates the baseline period, and dark grey shading indicates the target period. The figure compares ERA5, the CMIP6 ensemble median, and individual models, showing trends for ERA5 and the ensemble median, as well as the ensemble spread and slope values of the trends.
3.6. Results summary#
ERA5 indicates a general intensification of drought conditions over the Mediterranean region when comparing the baseline (1961–1990) and target (1991–2020) periods.
The CMIP6 ensemble median captures this SPEI6 change signal, but the magnitude of the increase in drought conditions is underestimated, particularly over eastern Spain, southern France, and much of Turkey.
With respect to inter-model spread, models do not reproduce the same spatial pattern. The largest spread appears over North African countries bordering the Mediterranean, Turkey, Lebanon, and Syria, and to a lesser extent over the Balkans, Portugal, and central and western Spain.
Time series of spatial means over the Mediterranean confirm this increase in drought conditions and the underestimation by CMIP6 models, with trends of –0.15 for ERA5 and –0.08 (about half) for the CMIP6 median.
3.7. Implications for the users#
Models do not reproduce the same exact spatial pattern, but the ensemble median captures the SPEI6 change. This underlines the importance of considering large ensembles rather than relying on a single or a few models.
Although the CMIP6 ensemble median captures the intensification of drought conditions, caution is needed when looking into the future. Precipitation projections generally involve more uncertainty than temperature, and model biases may evolve under changing climate conditions [9].
This assessment shows consistent results with those obtained in the C3S Atlas. In addition, it provides users with insight into the rationale behind SPEI and how models reproduce historical conditions over the Mediterranean, reinforcing its value as a tool for water management and drought preparedness.
The pronounced inter-model spread over North Africa, Turkey, and the eastern Mediterranean implies that users in these regions may face higher future uncertainty. This highlights the need to stress-test adaptation strategies against a wide range of plausible futures rather than relying on a single scenario.
ℹ️ If you want to know more#
Key resources#
Some key resources and further reading were linked throughout this assessment.
The CDS catalogue entries for the data used were:
CMIP6 climate projections (daily - daily maximum near-surface air temperature, daily minimum near-surface air temperature and precipitation): https://cds.climate.copernicus.eu/datasets/projections-cmip6?tab=overview
ERA5 hourly data on single levels from 1940 to present (2m temperature and total precipitation): https://cds.climate.copernicus.eu/datasets/reanalysis-era5-pressure-levels-monthly-means?tab=overview
Code libraries used:
C3S EQC custom functions,
c3s_eqc_automatic_quality_control, prepared by B-Open
References#
[1] Vicente-Serrano, S. M., Beguerı́a, S., and López-Moreno, J. I., 2010. A multiscalar drought index sensitive to global warming: The standardized precipitation evapotranspiration index, Journal of Climate, 23, 1696–1718. https://doi.org/10.1175/2009JCLI2909.1
[2] Wilhite, D. A., 2000. Droughts as a natural hazard: concepts and definitions. In: DROUGHT, A Global Assessment, vol. I and II, Routledge Hazards and Disasters Series, Routledge.
[3] Vicente-Serrano, S. M., Beguerı́a, S., Lorenzo-Lacruz, J., Camarero, J. J., López-Moreno, J. I., Azorin-Molina, C., Revuelto, J., Morán-Tejeda, E., and Sanchez-Lorenzo, A., 2012. Performance of drought indices for ecological, agricultural, and hydrological applications, Earth Interactions, 16, https://doi.org/10.1175/2012EI000434.1
[4] Yao, N., Li, L., Feng, P., Feng, H., Liu, D. L., Liu, Y., Jiang, K., Hu, X., and Li, Y., 2020. Projections of drought characteristics in China based on a standardized precipitation and evapotranspiration index and multiple GCMs. Science of The Total Environment, 704, 135245. https://doi.org/10.1016/j.scitotenv.2019.135245
[5] Tuong, V. Q., Kiet, B. A., and Pham, T. T., 2025. Assessment of Future Drought Characteristics Using Various Temporal Scales and Multiple Drought Indices over Mekong Basin Under Climate Changes. Water, 17(10), 1507. https://doi.org/10.3390/w17101507
[6] Iturbide, M., Gutiérrez, J. M., Alves, L. M., Bedia, J., Cerezo-Mota, R., Cimadevilla, E., Cofiño, A. S., Di Luca, A., Faria, S. H., Gorodetskaya, I. V., Hauser, M., Herrera, S., Hennessy, K., Hewitt, H. T., Jones, R. G., Krakovska, S., Manzanas, R., Martínez-Castro, D., Narisma, G. T., Nurhati, I. S., Pinto, I., Seneviratne, S. I., van den Hurk, B., and Vera, C. S., 2020. An update of IPCC climate reference regions for subcontinental analysis of climate model data: definition and aggregated datasets, Earth Syst. Sci. Data, 12, 2959–2970, https://doi.org/10.5194/essd-12-2959-2020
[7] Hargreaves, G. H., and Samani, Z. A., 1985. Reference crop evapotranspiration from temperature, Applied engineering in agriculture, pp. 96–99. https://doi.org/10.13031/2013.26773
[8] Beguerı́a, S., Vicente-Serrano, S. M., Reig, F., and Latorre, B., 2014. Standardized precipitation evapotranspiration index (spei) revisited: Parameter fitting, evapotranspiration models, tools, datasets and drought monitoring, International Journal of Climatology, 34, 3001–3023. https://doi.org/10.1002/joc.3887
[9] Schmith, T., Thejll, P., Berg, P., Boberg, F., Christensen, O. B., Christiansen, B., Christensen, J. H., Madsen, M. S., and Steger, C., 2021. Identifying robust bias adjustment methods for European extreme precipitation in a multi-model pseudo-reality setting, Hydrol. Earth Syst. Sci., 25, 273–290, https://doi.org/10.5194/hess-25-273-2021
