
7.1. Consistency between the C3S Atlas dataset and its origins: Case study#
Production date: 2025-09-30.
Dataset version: 2.0.
Produced by: Olivier Burggraaff, Nicole Reynolds (National Physical Laboratory).
🌍 Use case: Retrieving climate indicators from the Copernicus Interactive Climate Atlas#
❓ Quality assessment question#
Are the climate indicators in the dataset underpinning the Copernicus Interactive Climate Atlas consistent with their origin datasets?
Can the dataset underpinning the Copernicus Interactive Climate Atlas be reproduced from its origin datasets?
The Copernicus Interactive Climate Atlas, or C3S Atlas for short, is a C3S web application providing an easy-to-access tool for exploring climate projections, reanalyses, and observational data [Gutiérrez+24]. Version 2.0 of the application allows the user to interact with 12 datasets:
Type |
Dataset |
|---|---|
Climate Projection |
CMIP6 |
Climate Projection |
CMIP5 |
Climate Projection |
CORDEX-CORE |
Climate Projection |
CORDEX-EUR-11 |
Reanalysis |
ERA5 |
Reanalysis |
ERA5-Land |
Reanalysis |
ORAS5 |
Reanalysis |
CERRA |
Observations |
E-OBS |
Observations |
BERKEARTH |
Observations |
CPC |
Observations |
SST-CCI |
These datasets are provided through an intermediary dataset, the Gridded dataset underpinning the Copernicus Interactive Climate Atlas or C3S Atlas dataset for short [C3S Atlas dataset]. Compared to their origins, the versions of the climate datasets within the C3S Atlas dataset have been processed following the workflow in Figure 7.1.1.

Fig. 7.1.1 Schematic representation of the workflow for the production of the C3S Atlas dataset from its origin datasets, from the User-tools for the C3S Atlas.#
Because a wide range of users interact with climate data through the C3S Atlas application, it is crucial that the underpinning dataset represent its origins correctly. In other words, the C3S Atlas dataset must be consistent with and reproducible from its origins. Here, we assess this consistency and reproducibility by comparing climate indicators retrieved from the C3S Atlas dataset with their equivalents calculated from the origin dataset, mirroring the workflow from Figure 7.1.1. While a full analysis and reproduction of every record within the C3S Atlas dataset is outside the scope of quality assessment (and would require high-performance computing infrastructure), a case study with a narrower scope probes these quality attributes of the dataset and can be a jumping-off point for further analysis by the reader.
This notebook is part of a series:
Notebook |
Contents |
|---|---|
Consistency between the C3S Atlas dataset and its origins: Case study |
Comparison between C3S Atlas dataset and one origin dataset (CMIP6) for one indicator ( |
Consistency between the C3S Atlas dataset and its origins: Multiple indicators |
Comparison between C3S Atlas dataset and one origin dataset (CMIP6) for multiple indicators. |
Consistency between the C3S Atlas dataset and its origins: Multiple origin datasets |
Comparison between C3S Atlas dataset and multiple origin datasets for one indicator. |
📢 Quality assessment statement#
These are the key outcomes of this assessment
Climate indicators (here
tx35) provided by the C3S Atlas dataset are highly consistent with values calculated from its origin datasets (here the CMIP6 multi-model ensemble).Differences between the C3S Atlas dataset and a manual reproduction are rare (fewer than 0.1% of pixel pairs) and generally negligible (median absolute difference of 0).
The C3S Atlas is traceable and reproducible, and can be confidently used to view, analyse, and download climate data.
📋 Methodology#
This quality assessment tests the consistency between climate indicators retrieved from the Gridded dataset underpinning the Copernicus Interactive Climate Atlas [C3S Atlas dataset] and their equivalents calculated from the origin datasets, as well as the reproducibility of said dataset.
This notebook starts the quality assessment with a simple case study: one climate indicator derived from one origin dataset.
This indicator is tx35,
the Monthly count of days with maximum near-surface (2-metre) air temperature above 35 °C,
derived from the CMIP6 multi-model ensemble [CMIP6 dataset].
This notebook mirrors one of the reproducibility demonstrations written by the data provider.
The analysis and results are organised in the following steps, which are detailed in the sections below:
Install User-tools for the C3S Atlas.
Import all required libraries.
Define indicator.
Define helper functions.
2. Calculate indicator from the origin dataset
Download data from the origin dataset.
Homogenise data.
Calculate indicator.
Regrid the origin data to the C3S Atlas grid.
3. Retrieve indicator from the C3S Atlas dataset
Download data from the C3S Atlas dataset.
Consistency: Compare the C3S Atlas and reproduced datasets on native grids.
Reproducibility: Compare the C3S Atlas and reproduced datasets on the C3S Atlas grid.
📈 Analysis and results#
1. Code setup#
Note
This notebook uses earthkit for downloading (earthkit-data) and visualising (earthkit-plots) data. Because earthkit is in active development, some functionality may change after this notebook is published. If any part of the code stops functioning, please raise an issue on our GitHub repository so it can be fixed.
Install the User-tools for the C3S Atlas#
This notebook uses the User-tools for the C3S Atlas, which can be installed from GitHub using pip.
For convenience, the following cell can do this from within the notebook.
Further details and alternative options for installing this library are available in its documentation.
Import required libraries#
In this section, we import all the relevant packages needed for running the notebook.
Define indicators#
This section defines functions and variables for calculating and using the climate indicators:
The following cell defines earthkit-plots styles for the indicators. These styles define the colour maps and colour bar ranges for each quantity. Earthkit-plots styles are explained further in the corresponding documentation.
Helper functions#
This section defines some functions and variables used in the following analysis, allowing code cells in later sections to be shorter and ensuring consistency.
Data (pre-)processing#
The following functions handle the homogenisation of origin data to a consistent format using the User-tools for the C3S Atlas:
The following functions handle regridding data based on ESMF as implemented in the User-tools for the C3S Atlas. This step is explained in more detail in the relevant section below.
The following functions aid in sub-selecting data, e.g. selecting one model from the ensemble included in the C3S Atlas dataset or selecting data for a specific time frame:
Statistics#
The following functions calculate the difference (absolute / relative) between datasets, handling metadata etc.:
The following functions calculate and display metrics for the difference between two datasets, e.g. mean and median deviation:
Visualisation#
The following cells contain functions for plotting results, starting with some base helper functions (e.g. displaying in Jupyter Notebook or Jupyter Book style, adding textboxes with consistent formatting, etc.):
The following functions are also base helper functions, but specific to geospatial plots:
The following cell contains functions for geospatial comparisons between datasets on their native grids or on a common grid (the latter also showing the per-pixel difference):
The following cell contains functions for histogram comparisons between datasets on their native grids or on a common grid (the latter also showing the per-pixel difference):
2. Calculate indicator from the origin dataset#
Download data#
This assessment examines the dataset in two years (2060 and 2080),
for one ensemble member
and
for one climate scenario
– which is not how climate projection data are normally used.
Good practice in climate science is to look at multi-year statistics and trends,
across multiple ensemble members.
However, since the purpose of this assessment is to assess the consistency and reproducibility of the post-processing performed to produce the C3S Atlas dataset,
it is valid to use a subset of the data here.
The specific subset used can be easily tweaked by changing the EXPERIMENT, MODEL, and YEARS variables in this section.
This notebook uses earthkit-data to download files from the CDS. If you intend to run this notebook multiple times, it is highly recommended that you enable caching to prevent having to download the same files multiple times. If you prefer not to use earthkit, the following requests can also be used with the cdsapi module. In either case (earthkit-data or cdsapi), it is required to set up a CDS account and API key as explained on the CDS website.
The first step is to define the parameters that will be shared between the download of the origin dataset (here CMIP6) and the C3S Atlas dataset (in the next section), namely the experiment and model member.
Next, the request to download the corresponding data from CMIP6 is defined,
in this notebook choosing only the variable necessary to calculate the tx35 indicator, namely daily_maximum_near_surface_air_temperature.
Because the indicator will be calculated for multiple years, the year parameter of the request is left out for now.
This case study examines two years, namely 2060 and 2080, so there are two corresponding requests. This can easily be changed or extended to use different years. Separate requests are defined for each year, rather than one combined request, to limit the size per request.
Earthkit downloads the data as a field list.
This is converted into an xarray dataset, which can be used in the following analysis.
<xarray.Dataset> Size: 167MB
Dimensions: (time: 730, bnds: 2, lat: 192, lon: 288)
Coordinates:
* time (time) object 6kB 2060-01-01 12:00:00 ... 2080-12-31 12:00:00
* lat (lat) float64 2kB -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 2kB 0.0 1.25 2.5 3.75 ... 355.0 356.2 357.5 358.8
height float64 8B 2.0
Dimensions without coordinates: bnds
Data variables:
time_bnds (time, bnds) object 12kB dask.array<chunksize=(1, 2), meta=np.ndarray>
lat_bnds (time, lat, bnds) float64 2MB dask.array<chunksize=(365, 192, 2), meta=np.ndarray>
lon_bnds (time, lon, bnds) float64 3MB dask.array<chunksize=(365, 288, 2), meta=np.ndarray>
tasmax (time, lat, lon) float32 161MB dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
Attributes: (12/48)
Conventions: CF-1.7 CMIP-6.2
activity_id: ScenarioMIP
branch_method: standard
branch_time_in_child: 60225.0
branch_time_in_parent: 60225.0
comment: none
... ...
title: CMCC-ESM2 output prepared for CMIP6
variable_id: tasmax
variant_label: r1i1p1f1
license: CMIP6 model data produced by CMCC is licensed und...
cmor_version: 3.6.0
tracking_id: hdl:21.14100/a1513d6f-5325-4b99-bbe6-9b096557b100Homogenise data#
One of the steps in the C3S Atlas dataset production chain is homogenisation, i.e. ensuring consistency between data from different origin datasets.
This homogenisation is implemented in the User-tools for the C3S Atlas, specifically the c3s_atlas.fixers.apply_fixers function.
The following changes are applied:
The names of the spatial coordinates are standardised to
[lon, lat].Longitude is converted from
[0...360]to[-180...180]format.The time coordinate is standardised to the CF standard calendar.
Variable units are standardised (e.g. °C for temperature).
Variables are resampled / aggregated to the required temporal resolution.
The homogenisation is applied in the following code cell.
The apply_fixers function describes the different homogenisation steps as it applies them;
this can be read by expanding the following cell outputs.
<xarray.Dataset> Size: 162MB
Dimensions: (time: 732, lat: 192, lon: 288)
Coordinates:
* lat (lat) float64 2kB -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 2kB -178.8 -177.5 -176.2 -175.0 ... 177.5 178.8 180.0
* time (time) datetime64[ns] 6kB 2060-01-01 2060-01-02 ... 2080-12-31
height float64 8B 2.0
Data variables:
tasmax (time, lat, lon) float32 162MB dask.array<chunksize=(1, 192, 288), meta=np.ndarray>
Attributes: (12/48)
Conventions: CF-1.7 CMIP-6.2
activity_id: ScenarioMIP
branch_method: standard
branch_time_in_child: 60225.0
branch_time_in_parent: 60225.0
comment: none
... ...
title: CMCC-ESM2 output prepared for CMIP6
variable_id: tasmax
variant_label: r1i1p1f1
license: CMIP6 model data produced by CMCC is licensed und...
cmor_version: 3.6.0
tracking_id: hdl:21.14100/a1513d6f-5325-4b99-bbe6-9b096557b100Calculate indicator#
The climate indicator,
in this notebook only tx35,
is calculated using xclim.
The function cal_tx35, defined above, performs the calculation.
This function needs to be applied year-by-year, which is achieved using groupby.
<xarray.Dataset> Size: 11MB
Dimensions: (time: 24, lat: 192, lon: 288)
Coordinates:
* lat (lat) float64 2kB -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
* lon (lon) float64 2kB -178.8 -177.5 -176.2 -175.0 ... 177.5 178.8 180.0
height float64 8B 2.0
* time (time) datetime64[ns] 192B 2060-01-01 2060-02-01 ... 2080-12-01
Data variables:
tx35 (time, lat, lon) int64 11MB dask.array<chunksize=(1, 192, 288), meta=np.ndarray>Regrid to C3S Atlas grid#
Note
This notebook uses xESMF for regridding data. xESMF is most easily installed using mamba/conda as explained in its documentation. Users who cannot or do not wish to use mamba/conda can manually compile and install ESMF on their machines. In future, this notebook will use earthkit-regrid instead, once it reaches suitable maturity.
The final step in the processing is regridding to the standardised grid used in the C3S Atlas dataset (Figure 7.1.1).
This is performed through a custom function in the User-tools for the C3S Atlas,
specifically c3s_atlas.interpolation.
This function is based on ESMF, as noted above.
Note that the C3S Atlas workflow calculates indicators first, then regrids. For operations that involve averaging, like smoothing and regridding, the order of operations can affect the result, especially in areas with steep gradients [Burggraaff+20]. Examples of such areas for a temperature index are coastlines and mountain ranges. In the case of C3S Atlas, this order of operations was a conscious choice to preserve the “raw” signals, e.g. preventing extreme temperatures from being smoothed out. However, it can affect the indicator values and therefore must be considered when using the C3S Atlas application or dataset.
<xarray.Dataset> Size: 12MB
Dimensions: (lon: 360, lat: 180, time: 24, bnds: 2)
Coordinates:
* lon (lon) float64 3kB -179.5 -178.5 -177.5 ... 177.5 178.5 179.5
* lat (lat) float64 1kB -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
* time (time) datetime64[ns] 192B 2060-01-01 2060-02-01 ... 2080-12-01
Dimensions without coordinates: bnds
Data variables:
tx35 (time, lat, lon) int64 12MB dask.array<chunksize=(1, 180, 360), meta=np.ndarray>
lon_bnds (lon, bnds) float64 6kB -180.0 -179.0 -179.0 ... 179.0 179.0 180.0
lat_bnds (lat, bnds) float64 3kB -90.0 -89.0 -89.0 -88.0 ... 89.0 89.0 90.0
crs int64 8B 0
height float64 8B 2.03. Retrieve indicator from the C3S Atlas dataset#
Here, we download the same indicator as above directly from the Gridded dataset underpinning the Copernicus Interactive Climate Atlas so the values can be compared.
The C3S Atlas dataset provides all years and members at the same time, so for convenience we pull out only the relevant entries:
<xarray.Dataset> Size: 6MB
Dimensions: (lat: 180, bnds: 2, lon: 360, time: 24)
Coordinates:
* lat (lat) float64 1kB -89.5 -88.5 -87.5 ... 87.5 88.5 89.5
* lon (lon) float64 3kB -179.5 -178.5 -177.5 ... 178.5 179.5
* time (time) datetime64[ns] 192B 2060-01-01 ... 2080-12-01
member_id <U45 180B dask.array<chunksize=(), meta=np.ndarray>
gcm_institution <U19 76B dask.array<chunksize=(), meta=np.ndarray>
gcm_model <U16 64B dask.array<chunksize=(), meta=np.ndarray>
gcm_variant <U8 32B dask.array<chunksize=(), meta=np.ndarray>
threshold35c float64 8B ...
height2m float64 8B ...
Dimensions without coordinates: bnds
Data variables:
lat_bnds (lat, bnds) float64 3kB dask.array<chunksize=(180, 2), meta=np.ndarray>
lon_bnds (lon, bnds) float64 6kB dask.array<chunksize=(360, 2), meta=np.ndarray>
time_bnds (time, bnds) datetime64[ns] 384B dask.array<chunksize=(24, 2), meta=np.ndarray>
tx35 (time, lat, lon) float32 6MB dask.array<chunksize=(24, 26, 52), meta=np.ndarray>
crs int32 4B ...
Attributes: (12/26)
Conventions: CF-1.9 ACDD-1.3
title: Copernicus Interactive Climate Atlas: gridded...
summary: Monthly/annual gridded data from observations...
institution: Copernicus Climate Change Service (C3S)
producers: Institute of Physics of Cantabria (IFCA, CSIC...
license: CC-BY 4.0, https://creativecommons.org/licens...
... ...
geospatial_lon_min: -180.0
geospatial_lon_max: 180.0
geospatial_lon_resolution: 1.0
geospatial_lon_units: degrees_east
date_created: 2024-12-05 16:37:49.749769+01:00
tracking_id: 1a7a60e7-7787-48b5-b18f-a7bf7b4de4be4. Results#
This section contains the comparison between the indicator values retrieved from the C3S Atlas dataset vs those reproduced from the origin dataset.
The datasets are first compared on their native grids. This means a point-by-point comparison is not possible (because the points are not equivalent), but the distributions can be compared geospatially and overall. This qualitative comparison probes the consistency quality attribute: Are the climate indicators in the dataset underpinning the Copernicus Interactive Climate Atlas consistent with their origin datasets?
Second, the C3S Atlas dataset is compared to the indicators derived from the origin dataset and regridded to the C3S Atlas grid. This makes a quantitative point-by-point comparison possible. This second comparison probes how well the dataset underpinning the Copernicus Interactive Climate Atlas can be reproduced from its origin datasets, based on the workflow (Figure 7.1.1).
For the geospatial comparison,
we display the values of the indicator for one month,
across one region and globally.
For the example of tx35,
June
should provide many warm days in the Northern hemisphere with significant spatial variation.
As an example, we display the results across
Europe.
This region can easily be modified in the following code cell using the domains provided by earthkit-plots.
Some examples are provided in the cell (commented out using #).
Consistency: Comparison on native grids#
It is clear from Figures 7.1.2 and 7.1.3 that the C3S Atlas dataset and its reproduction from the origin display very similar patterns, as expected. The general distribution of indicator values (in both years) is the same, in this example showing more hot days in northern Africa, the Middle East, and eastern Europe than in western and northern Europe. Small differences appear in various places, likely the result of the regridding step in the C3S Atlas workflow as noted before. Larger differences can be seen in areas with steep temperature gradients, such as Italy with its mountains and coastlines. Figure 7.1.5 confirms that the overall distributions in indicator values (across all time and space dimensions) are very similar, but small differences exist.
For reference, Figure 7.1.4 displays the equivalent view in the C3S Atlas application. The application export is visually similar to Figure 7.1.3, showing the consistency between application and dataset, and hence showing that the conclusions from this quality assessment transfer to the C3S Atlas application. Differences are caused by the application displaying projection data as a multi-year average (here 2081–2100) as well as the difference in visualisation style.
Overall, we can conclude that the C3S Atlas dataset and its origins are highly consistent, but small differences exist due to the difference in grid. Users of the C3S Atlas dataset – and thus users of the C3S Atlas application – should be aware that the indicator values retrieved for a specific location may differ slightly from a manual analysis of the origin dataset. This effect may be larger or smaller for values aggregated over larger areas, such as the country and IPCC-AR6 region averages available in the C3S Atlas application, depending on the exact region and values used.
Lastly, it should be noted that the order of operations in the C3S Atlas workflow (regridding after the indicator calculation; Figure 7.1.1) results in values that may differ from the opposite order of operations (regridding before indicator calculation) [Burggraaff+20]. Assessing these differences is outside the scope of this assessment.
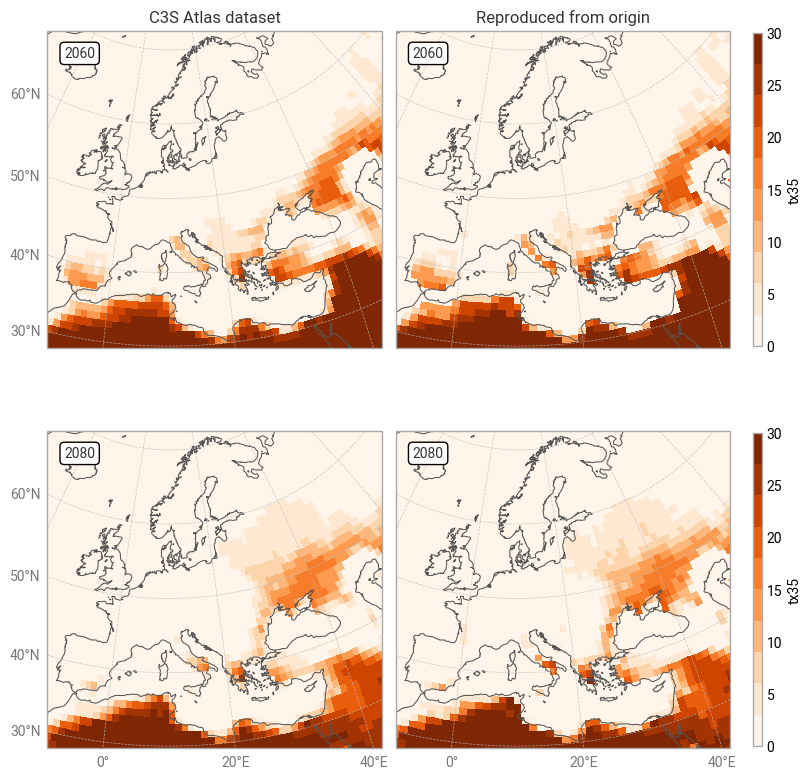
Fig. 7.1.2 Comparison between C3S Atlas dataset and reproduction for one indicator (tx35) in one month,
across Europe,
on the native grid of each dataset.#
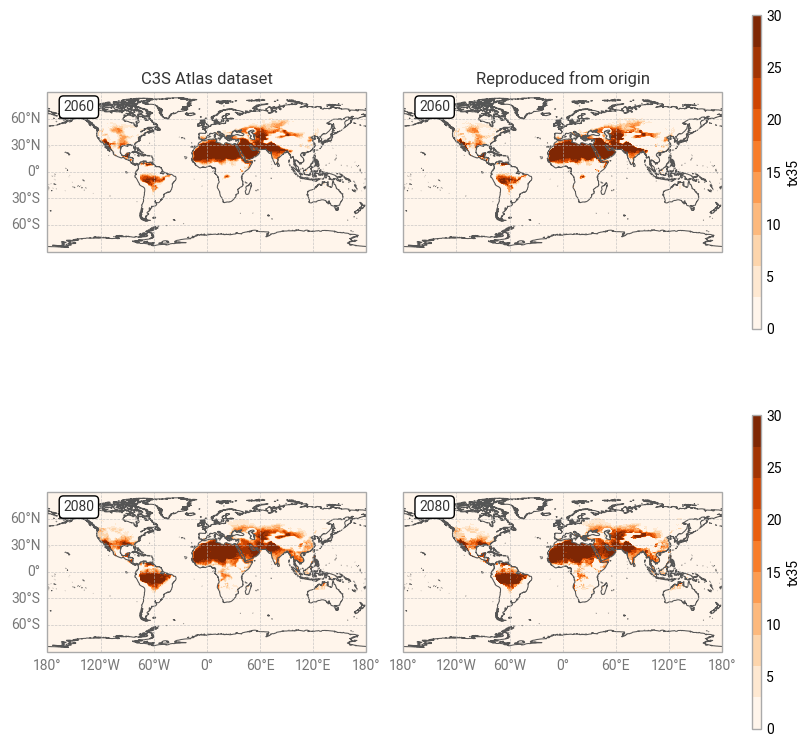
Fig. 7.1.3 Comparison between C3S Atlas dataset and reproduction for one indicator (tx35) in one month,
across the globe,
on the native grid of each dataset.#
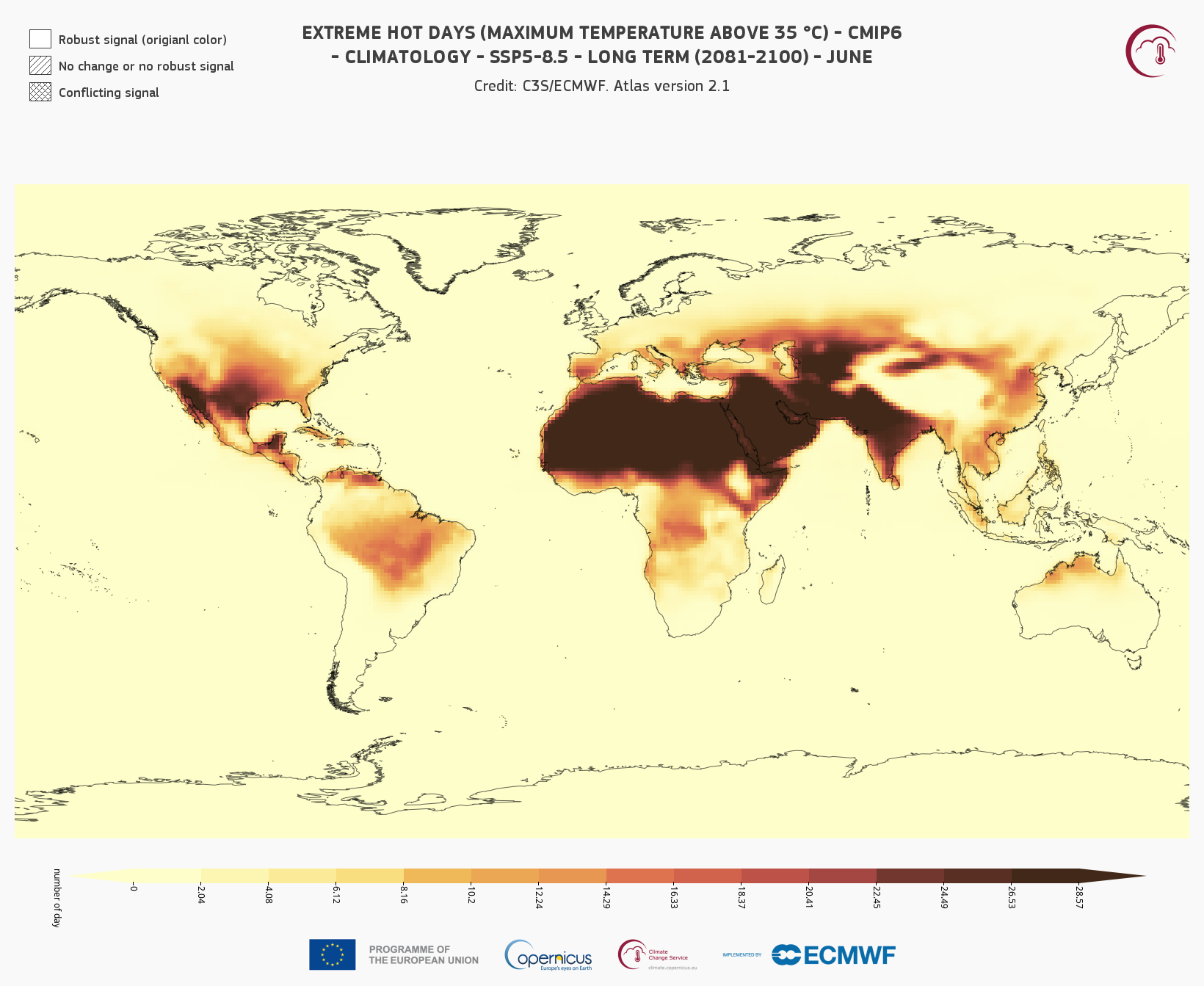
Fig. 7.1.4 C3S Atlas application view of tx35 indicator in June, averaged 2081–2100 in the SSP5-8.5 scenario,
for comparison with
Figure 7.1.3.#
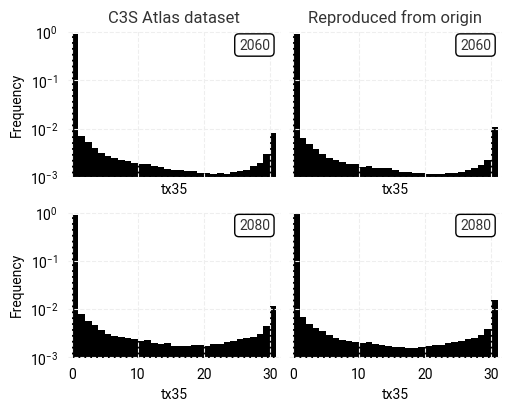
Fig. 7.1.5 Comparison between overall distributions of indicator (tx35) values in the C3S Atlas dataset and its reproduction,
across all spatial and temporal dimensions,
on the native grid of each dataset.#
Reproducibility: Comparison on C3S Atlas grid#
After regridding to the C3S Atlas grid, the indicator values reproduced from the origin dataset can be compared point-by-point to the values retrieved from the C3S Atlas dataset. We first examine some metrics that describe the difference Δ between corresponding pixels:
| Mean Δ | Median Δ | Median |Δ| | % where |Δ| ≥ ε | Median |Δ| [%] | Pearson r | |
|---|---|---|---|---|---|---|
| 2060 | -0.00077 | 0.00000 | 0.00000 | 0.07665 | 0.00000 | 0.99998 |
| 2080 | -0.00095 | 0.00000 | 0.00000 | 0.09388 | 0.00000 | 0.99998 |
It is clear that the two datasets are extremely close: the median difference and median absolute difference are both 0 and fewer than 0.1% of pixel pairs show a non-zero difference (defined here as |Δ| ≥ ε with ε = 10–5 to avoid floating-point errors). We can immediately conclude that the C3S Atlas dataset is highly reproducible from its origins using the provided workflow.
This conclusion is strengthened by examining the distributions of indicator values and their point-by-point differences (Figure 7.1.6). The vast majority of pixel pairs have a difference of 0, with very few ±1 and even fewer ±2 (note the logarithmic y-axis).
However, this raises an obvious question: why are there any differences at all? The geospatial distribution of differences (Figure 7.1.7) does not appear to correlate with any patterns in the indicator values themselves nor in geography. Since the differences are very small and seemingly random, a potential explanation is that they simply result from small changes in the origin dataset and/or the C3S Atlas workflow software between the production of the C3S Atlas dataset and the present day. Testing this hypothesis would require an extremely detailed analysis of all steps in the workflow, which is outside the scope of this assessment.
Setting this question affecting <0.1% of pixels aside,
we can confidently conclude
from the metrics and figures
that the C3S Atlas dataset is highly reproducible from its origins using the provided workflow,
at least for the tx35 indicator and CMIP6 dataset.
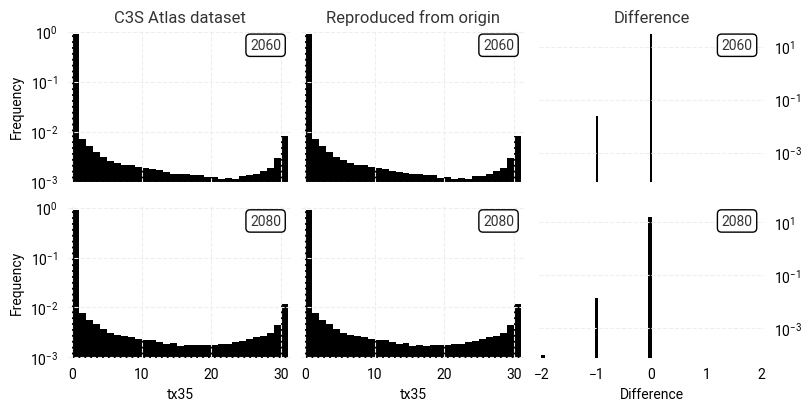
Fig. 7.1.6 Comparison between overall distributions of indicator (tx35) values in the C3S Atlas dataset and its reproduction
on the C3S Atlas grid,
across all spatial and temporal dimensions,
including the per-pixel difference.#
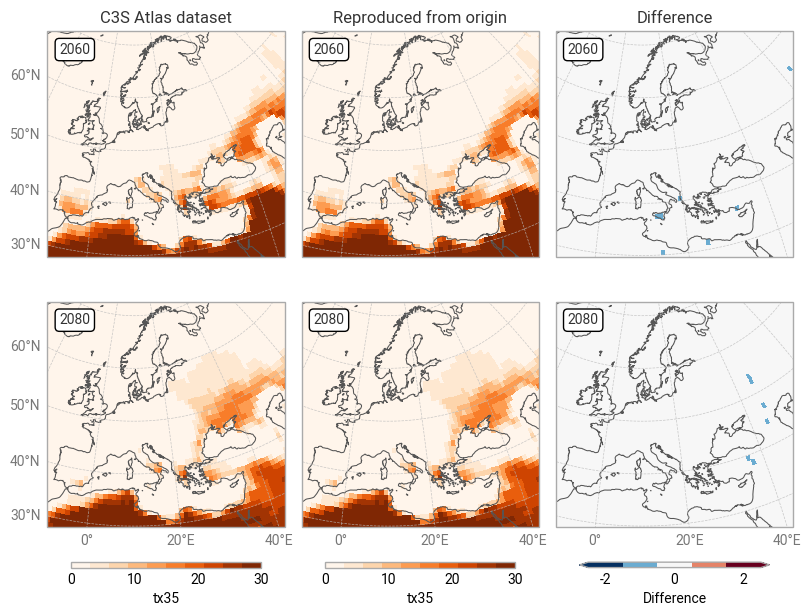
Fig. 7.1.7 Comparison between C3S Atlas dataset and reproduction for one indicator (tx35) in one month,
across Europe,
on the C3S Atlas dataset grid,
including the per-pixel difference.#
ℹ️ If you want to know more#
Key resources#
The CDS catalogue entries for the data used were:
Gridded dataset underpinning the Copernicus Interactive Climate Atlas: multi-origin-c3s-atlas
Consistency between the C3S Atlas dataset and its origins: Case study
Consistency between the C3S Atlas dataset and its origins: Multiple indicators
Consistency between the C3S Atlas dataset and its origins: Multiple origin datasets
CMIP6 climate projections: projections-cmip6
Code libraries used:
xclim climate indicator tools
More about the Copernicus Interactive Climate Atlas and its IPCC predecessor:
References#
[Gutiérrez+24] J. M. Gutiérrez et al., ‘The Copernicus Interactive Climate Atlas: a tool to explore regional climate change’, ECMWF Newsletter, vol. 181, pp. 38–45, Oct. 2024, doi: 10.21957/ah52ufc369.
[C3S Atlas dataset] Copernicus Climate Change Service, ‘Gridded dataset underpinning the Copernicus Interactive Climate Atlas’. Copernicus Climate Change Service (C3S) Climate Data Store (CDS), Jun. 17, 2024. doi: 10.24381/cds.h35hb680.
[CMIP6 dataset] Copernicus Climate Change Service, ‘CMIP6 climate projections’. Copernicus Climate Change Service (C3S) Climate Data Store (CDS), Mar. 23, 2021. doi: 10.24381/cds.c866074c.
[Burggraaff+20] O. Burggraaff, ‘Biases from incorrect reflectance convolution’, Optics Express, vol. 28, no. 9, pp. 13801–13816, Apr. 2020, doi: 10.1364/OE.391470.
