2.6. Seasonal stripes#
This Jupyter Notebook reproduces the Seasonal climate strips product from the C3S Atlas.
The seasonal stripe plot is like the climate stripes panel but including monthly values vertically instead of model results, displaying results for the ensemble multi-model median.
Note that climate stripes can be used for variables other than temperature to detect climate signals in the ensemble over time. In the case of the current notebook, precipitation is used
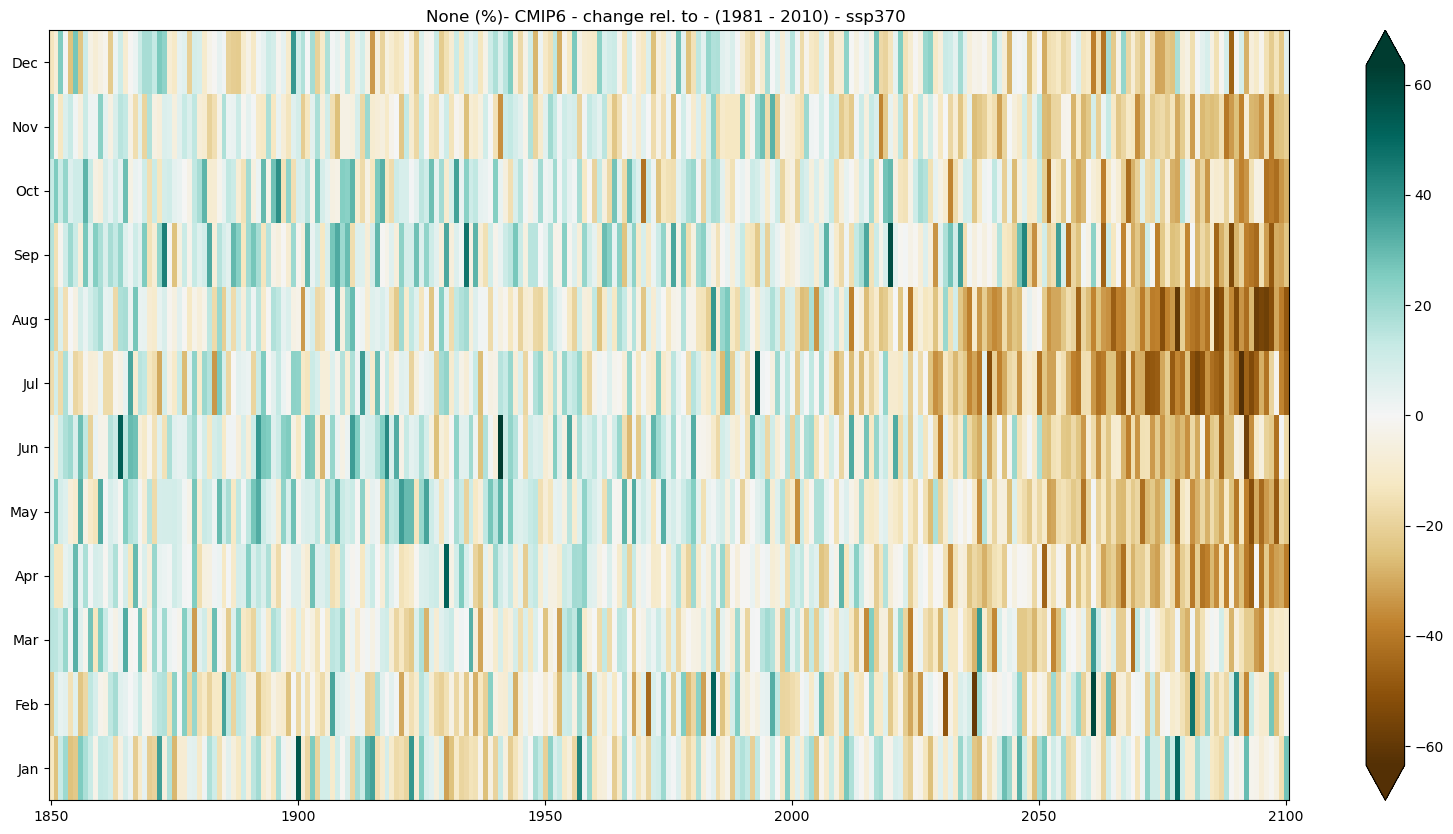
The figure below represents the seasonal climate stripe of precipitation for CMIP6 projections. It can be visualized in the C3S Atlas using the following Permalink
2.6.1. Load Python packages and clone and install the c3s-atlas GitHub repository from the ecmwf-projects#
Clone (git clone) the c3s-atlas repository and install them (pip install -e .).
Further details on how to clone and install the repository are available in the requirements section
import os
import xarray as xr
import glob
from datetime import date
import numpy as np
from pathlib import Path
import cdsapi
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
from c3s_atlas.utils import (
season_get_name,
extract_zip_and_delete,
)
from c3s_atlas.customized_regions import (
Mask
)
from c3s_atlas.analysis import (
seasonal_stripes,
)
from c3s_atlas.products import (
seasonal_stripe_plot,
)
from c3s_atlas.GWLs import (
load_GWLs,
select_member_GWLs,
get_selected_data
)
2.6.2. Download climate data with the CDS API#
To reduce data size and download time, a geographical subset focusing on a specific area within the European region (Spain) is selected.
cdsapi_url= "https://cds.climate.copernicus.eu/api"
cdsapi_key= ""
c = cdsapi.Client(url=cdsapi_url, key=cdsapi_key)
⚠️ Warning: Exposed API Credentials
For security reasons, it is not recommended to hardcode your Copernicus Climate Data Store (CDS) API credentials — such as
cdsapi_urlandcdsapi_key— directly in notebooks.Instead, it is best to store them securely in a
.cdsapircfile located in your home directory.📄 More info: CDS API - How to use the API
project = "CMIP6"
scenario = "ssp370"
var = 'r'
# directory to download the files
dest = Path('./data/CMIP6')
os.makedirs(dest, exist_ok=True)
Download historical data#
filename = 'r_CMIP6_historical_mon_185001-201412.zip'
dataset = "multi-origin-c3s-atlas"
request = {
"origin": "cmip6",
"experiment": "historical",
"period": "1850-2014",
"variable": "monthly_precipitation",
"bias_adjustment": "no_bias_adjustment",
'area': [44.5, -9.5, 35.5, 3.5]
}
c.retrieve(dataset, request).download(dest / filename)
extract_zip_and_delete(dest / filename)
Download SSP scenario#
filename = 'r_CMIP6_ssp370_mon_201501-210012.zip'
dataset = "multi-origin-c3s-atlas"
request = {
"origin": "cmip6",
"experiment": "ssp3_7_0",
"period": "2015-2100",
"variable": "monthly_precipitation",
"bias_adjustment": "no_bias_adjustment",
'area': [44.5, -9.5, 35.5, 3.5]
}
c.retrieve(dataset, request).download(dest / filename)
extract_zip_and_delete(dest / filename)
Concatenate historical and SSP scenarios#
Note that the historical and SSP scenarios may have a different number of members. Here, common members from the historical and SSP scenarios are concatenated into a single xarray.Dataset to facilitate their use going forward.
ds_hist = xr.open_dataset(dest / "r_CMIP6_historical_mon_185001-201412.nc")
ds_sce = xr.open_dataset(dest / "pr_CMIP6_ssp370_mon_201501-210012.nc")
mem_inters = np.intersect1d(ds_hist.member_id.values, ds_sce.member_id.values)
ds_hist = ds_hist.isel(member = np.isin(ds_hist.member_id.values, mem_inters))
ds_sce = ds_sce.isel(member = np.isin(ds_sce.member_id.values, mem_inters))
ds = xr.concat([ds_hist, ds_sce], dim = 'time')
attrs = {
"project" : project,
"scenario": scenario,
"variable": var,
"actual_year": date.today().year,
"unit" : ds[var].units
}
Select Region#
The Spanish region is used as an example from the predefined European countries regions to visualize the products. See customized_regions.ipynb notebook for more options and information.
mask = Mask(ds).European_contries(['ESP'])
filtered_ds = ds.where(mask)
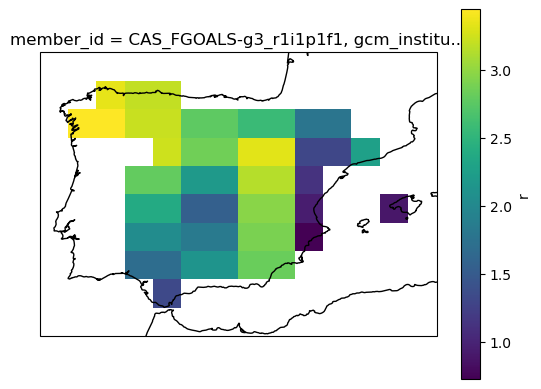
# mean spatial map for one member
ax = plt.axes(projection=ccrs.PlateCarree()) # Use PlateCarree for lat/lon projections
filtered_ds[var].isel(member=0).mean(dim='time').plot(ax=ax, transform=ccrs.PlateCarree())
ax.coastlines()
plt.show()
2.6.3. Analysis for the product#
The seasonal_stripes funtion reshapes the dataset into a matrix with months and years as dimensions.
seasonal_stripes_ds = seasonal_stripes(filtered_ds, var, project)
2.6.4. Plot#
The seasonal_stripe_plot function lets you select the colorbar (cmap) used to represent the stripes. Sequential colorbars are recommended for displaying absolute values, while diverging colorbars are better suited for visualizing changes or differences, as they effectively highlight variations around a central value.
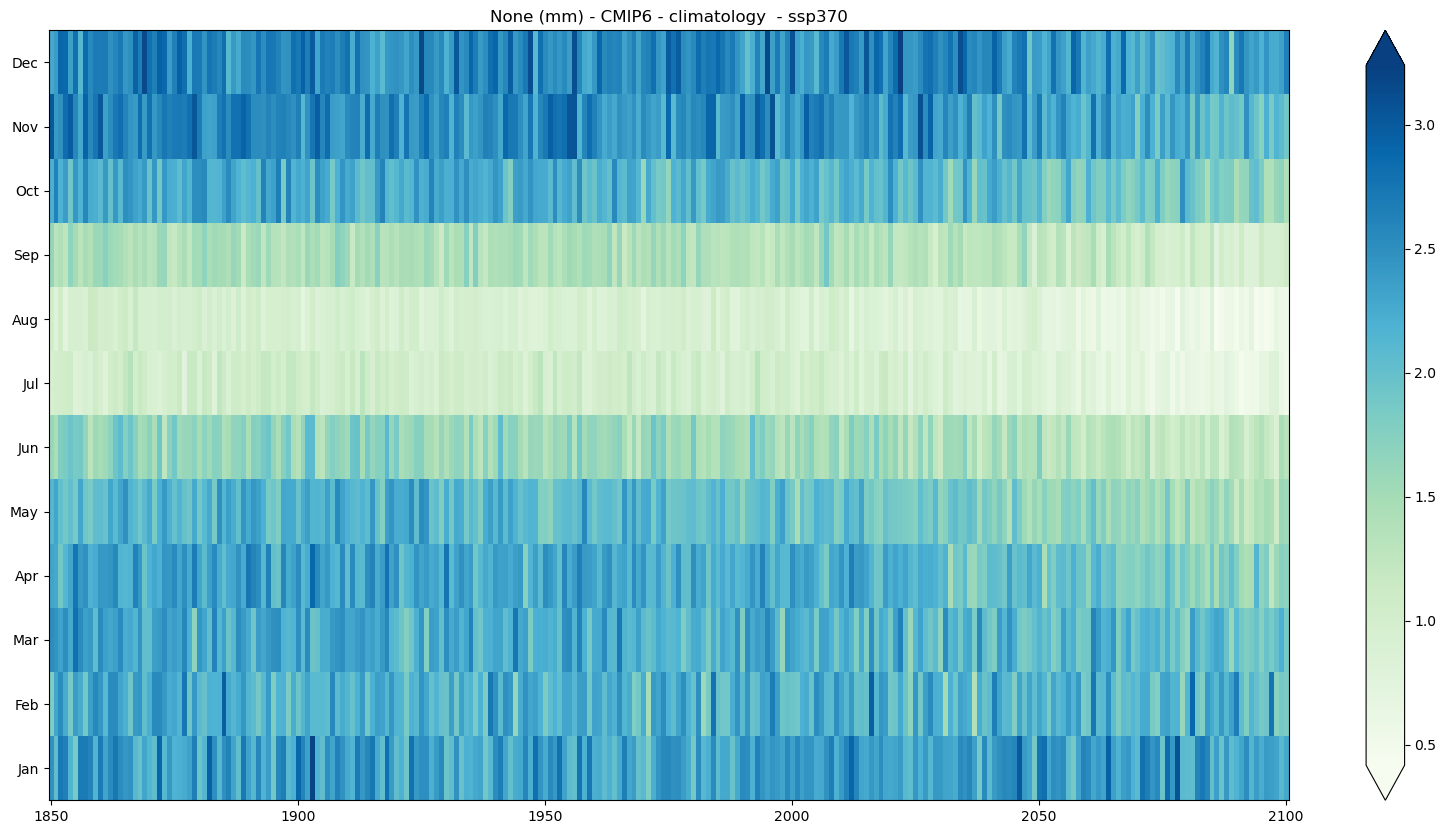
a) Climatology#
mode = 'climatology'
seasonal_stripe_fig = seasonal_stripe_plot(seasonal_stripes_ds, var,
attrs, mode = mode, cmap = 'GnBu')
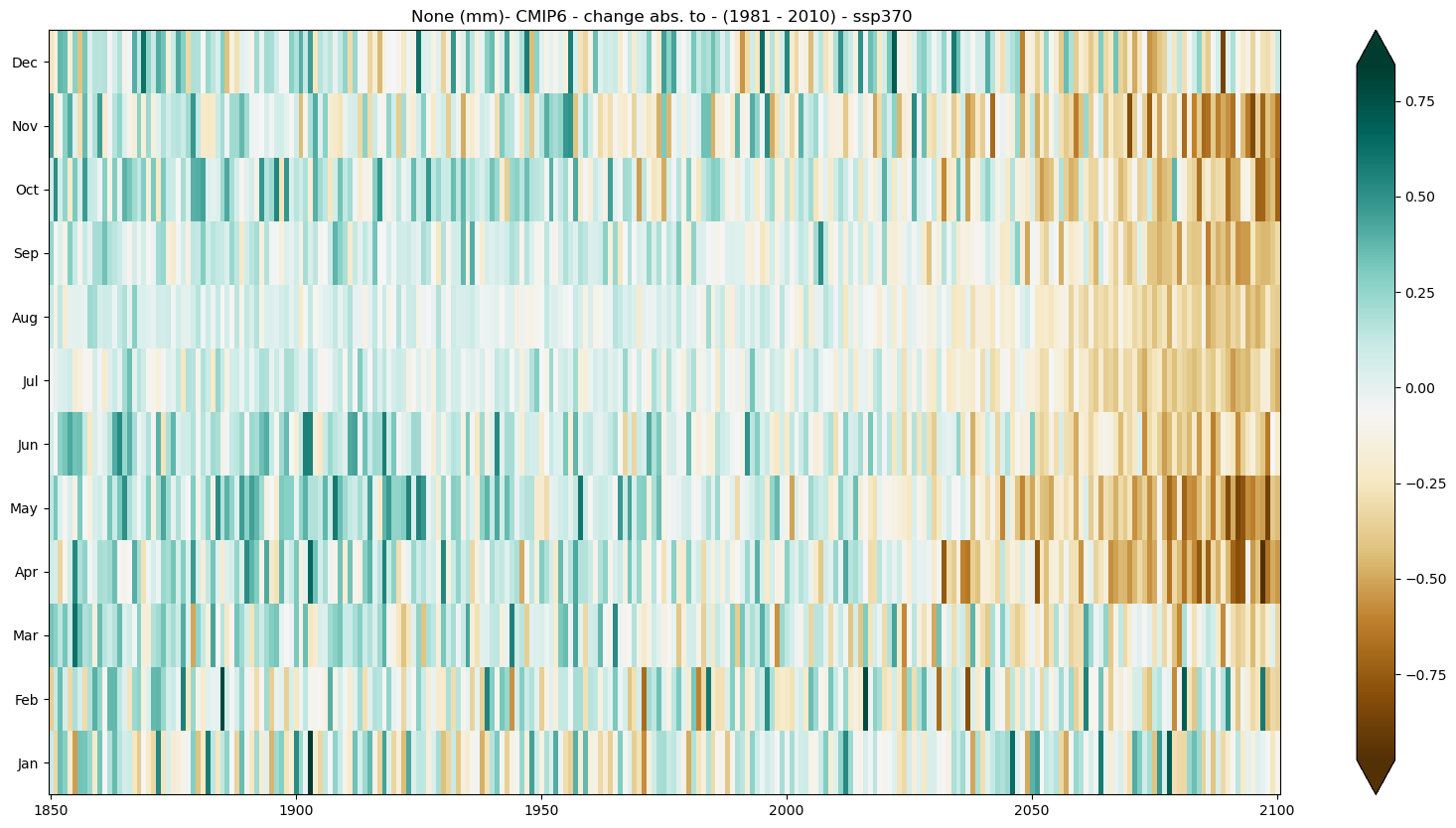
b) Absolute change#
mode = 'change'
diff = 'abs'
period=slice(1850, 2100)
baseline_period= slice(1981,2010)
seasonal_stripe_fig = seasonal_stripe_plot(seasonal_stripes_ds, var, attrs,
mode = mode, diff = diff,
period = period,
baseline_period = baseline_period,
cmap = 'BrBG')
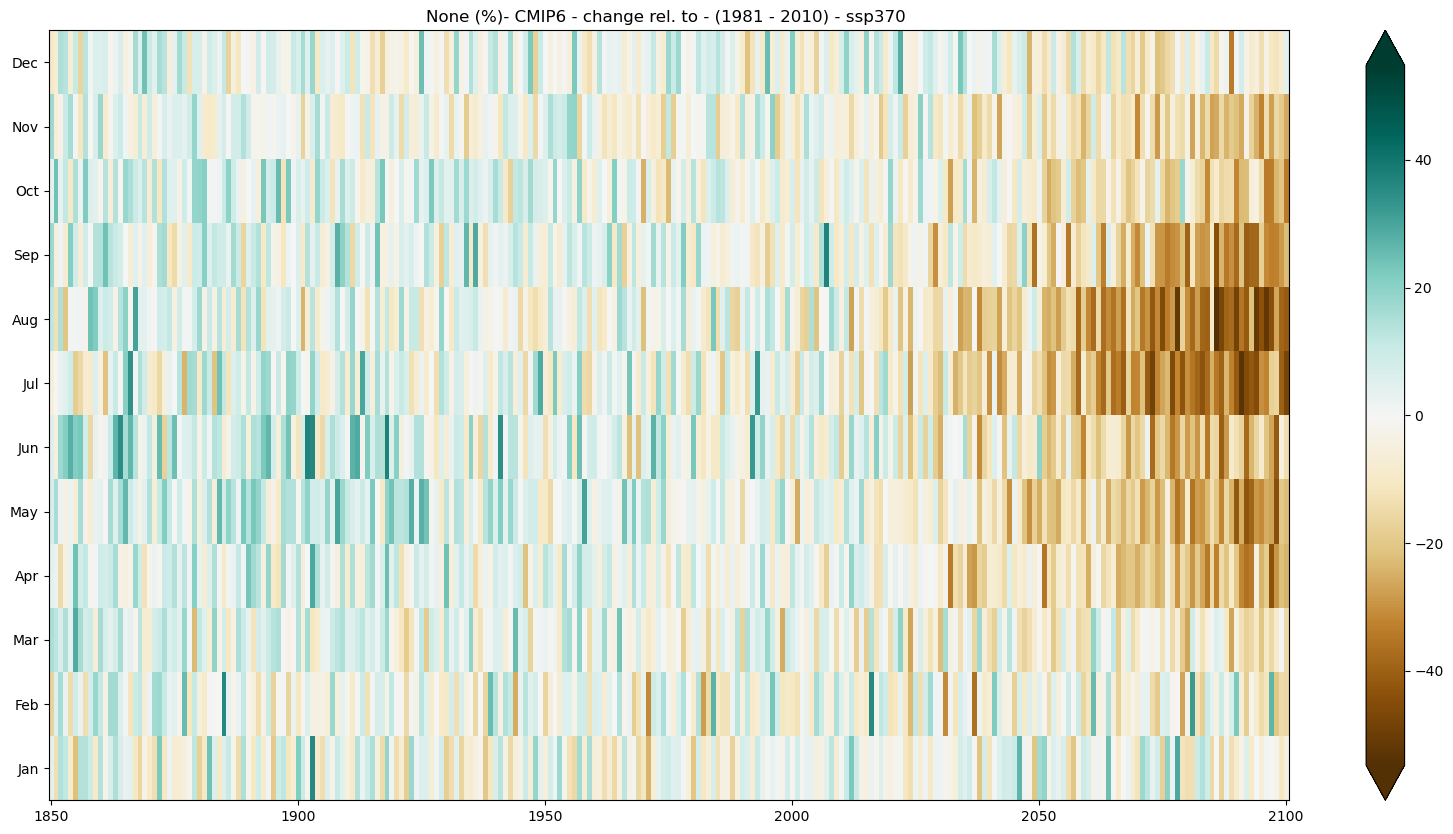
c) Relative change#
mode = 'change'
diff = 'rel'
period=slice(1850, 2100)
baseline_period = slice(1981,2010)
seasonal_stripe_fig = seasonal_stripe_plot(seasonal_stripes_ds,var, attrs,
mode = mode, diff = diff,
period = period,
baseline_period = baseline_period,
cmap = 'BrBG')
2.6.5. Global Warming Levels#
Here, the seasonal stripe is displayed for a specific Global Warming Level (GWL). To achieve this, the 20-year period in which each ensemble member reaches the chosen GWL is selected. The seasonal stripe is then shown, representing either climatology or change for this specific period and region (Spain in this notebook).
These periods are calculated in the notebook GWLs.ipynb for CMIP5 and CMIP6. For CORDEX, the results from the driving CMIP5 models are used.
GWL = '4'
#Load the data and get the intersection of the members
GWLs_ds = load_GWLs(project)
GWLs_members_with_period = select_member_GWLs(filtered_ds, GWLs_ds, project, scenario, GWL)
filtered_GWLs_ds = get_selected_data(filtered_ds, GWLs_members_with_period)
seasonal_stripes_GWLs_ds = seasonal_stripes(filtered_GWLs_ds, var, project)
mode = 'change'
diff = 'rel'
baseline_period = slice(1981,2010)
seasonal_stripe_fig = seasonal_stripe_plot(seasonal_stripes_GWLs_ds,var,
attrs, mode = mode, diff = diff,
baseline_period = baseline_period,
cmap = 'BrBG')